The research focus of the MTERMS Lab centers on harnessing artificial intelligence (AI) and informatics technologies—including natural language processing (NLP) and machine learning (ML)—to improve clinical decision support and advance knowledge discovery. Initially developed to extract and standardize medication information from clinical notes for real-time medication reconciliation in an electronic health record (EHR) system, MTERMS (Medical Text Extraction, Reasoning, and Mapping System) has evolved into a versatile platform and toolset supporting a wide range of clinical informatics applications and research projects.
The MTERMS ecosystem combines methods from linguistics, statistics, machine learning, and deep learning, applied to diverse biomedical data sources. Its functionality encompasses, but is not limited to, information extraction, retrieval, and encoding; data mining and knowledge discovery; document classification; relation identification; and predictive modeling. The MTERMS Lab employs MTERMS, alongside other NLP/ML technologies and generative large language models (LLMs), to address practical clinical challenges.
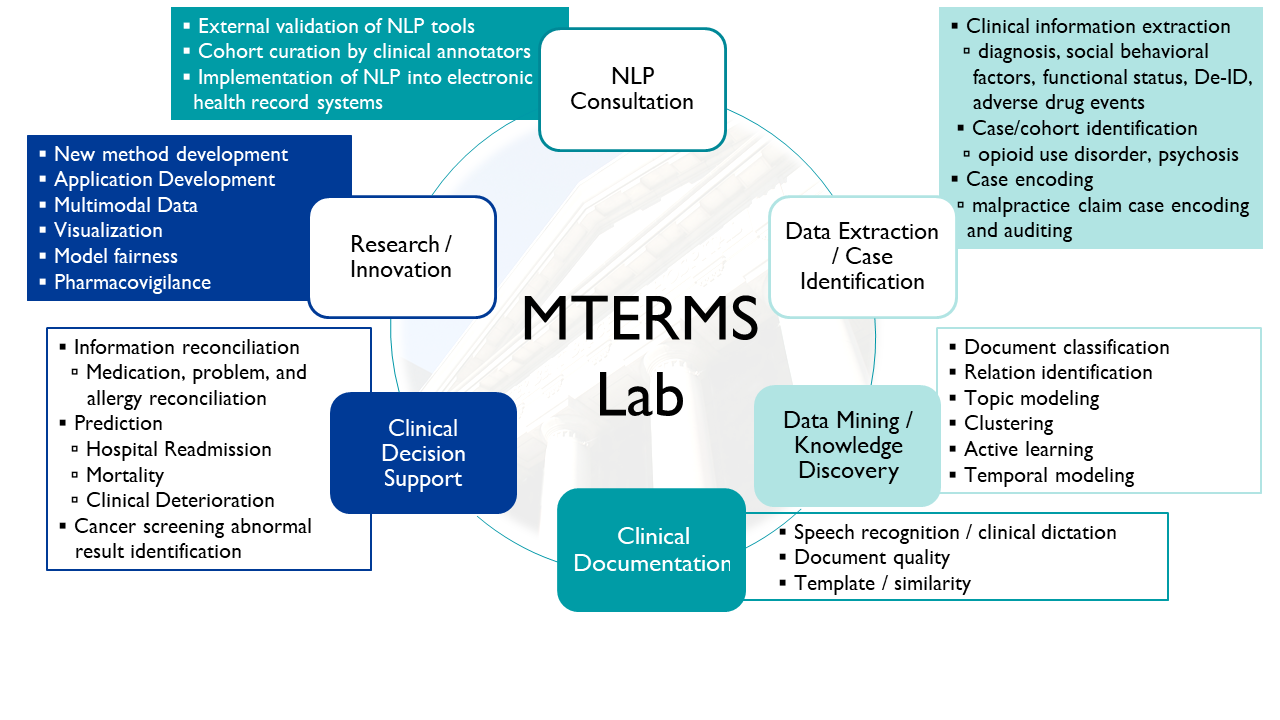
MTERMS has been successfully implemented across various clinical tasks, including clinical decision support, care coordination, malpractice coding and auditing, phenotyping, cohort identification, medication and allergy reconciliation, documentation quality and accuracy assessments, clinical outcome prediction, and more. Notably, MTERMS has been integrated with Epic for real-time applications, some of which have been demonstrated in clinical trials focused on tasks such as allergy reconciliation, detecting clinical deterioration, and identifying abnormal cancer screening results. Their work has garnered multiple awards and has been featured in prominent media outlets, including The Wall Street Journal, The New York Times, Reuters, and NPR.
Each year, the MTERMS lab provides hands-on research opportunities for graduate students, research fellows, and trainees, allowing them to gain experience in biomedical informatics while completing capstone projects or thesis work. The lab welcomes research and development collaborations, particularly those involving artificial intelligence, NLP, generative LLMs, and other advanced technologies applied to clinical use cases, with a special focus on enhancing clinical decision support and patient safety.
Below are some examples of our research work:
- Clinical Documentation
- Automatic error checking of various clinical documents, including misspellings in typed notes and “real-word” errors in dictated notes
- Mining similar notes to help generate note templates
- Information Extraction/Retrieval and Encoding
- Extracting diverse clinical information (e.g., diagnoses, symptoms, medications, social-behavioral information) from free-text clinical documents
- Encoding extracted information using standard medical terminologies (e.g., SNOMED-CT, ICD-9/10, RxNorm)
- Handling contextual information (e.g., sections, negations, status/changes)
- Generating output in standard (e.g., HL7 document standards) or custom formats
- De-identification of clinical notes
- Data Mining and Knowledge Discovery
- Documentation classification, e.g., classifying malpractice claim descriptions according to allegation, contributing factors, severity, and responsible service
- Clustering, e.g., discovering food allergen cross-sensitivity
- Relation identification, e.g., temporal reasoning over medical events
- Distributional semantic analysis, e.g., topic modeling to generate themes from social media posts and clinical documents
- Active learning, e.g., using word embeddings to assist information extraction tasks
- Clinical Decision Support
- Creating an application to facilitate clinical information reconciliation in the electronic health record
- Developing NLP algorithms to identify abnormal cancer screening results from free-text clinical reports to facilitate timely follow-up
- Developing a prototype application for care coordination
- Developing a system for automatic malpractice case encoding and auditing
- Developing predictive models for identifying patients for early palliative care interventions
- Identifying patient cohorts (e.g., patients with heart failure)
- Research, Innovation and others
- Addressing missingness and bias in multimodal health data to enhance AI algorithms
- Developing applications to identify adverse drug events (e.g., adverse drug reactions to opioids)
- Participating in community shared tasks (e.g., classifying psychiatric symptom severity for the 2016 i2b2 NLP Shared Task)